As you saw from the DNA Sequencing section, it is now possible to figure out the exact sequence of a gene or even the entire genome. When the sequence is compared between people who have a specific disease to people who do not have any disease, then it is possible to see if any differences in the sequence contributed to the disease. This is how many genetic diseases are discovered. Also, somatic mutations (acquired mutations) can be detected by comparing the sequence of a normal cell to the sequence of DNA from a cancer cell. The obvious question that shows up – if a disease causing mutation is known, is it possible to change it back to the normal sequence? For example, the T mutation in Sickle Cell Anemia (SCA) as explained in the Genetics Section. If it can be changed to A, then sickle cell anemia can be treated or at least controlled effectively. This is now possible –and it has already been done. https://www.npr.org/sections/health-shots/2021/12/31/1067400512/first-sickle-cell-patient-treated-with-crispr-gene-editing-still-thriving
https://news.berkeley.edu/2021/03/30/fda-approves-first-test-of-crispr-to-correct-genetic-defect-causing-sickle-cell-disease/Let us look at how this is possible
Modifying the sequence of DNA (editing) has been attempted since the 1980s. In fact, mouse stem cells were modified precisely by researchers like Mario Capecchi Martin Evans and Oliver Smithies. In 2007, they shared the Nobel prize for their work: https://www.nature.com/collections/blbjmwqqmg#current
By 2013, several other technologies were developed and most centered around the ability of certain enzymes called nucleases to cut DNA. These include engineered meganucleases, Zinc finger nucleases (ZFNs), TAL effector nucleases (TALENs) and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR/Cas9) nucleases.
https://www.nih.gov/news-events/gene-editing-digital-press-kit
The most popular is CRISPR as it is easier and can be very precise with some new modifications made to the method. The two scientists (Emmanuelle Charpentier and Jennifer Doudna) who developed this method won the Nobel Prize in 2020 and you can read about the impact of this method here:
https://www.nobelprize.org/uploads/2020/10/popular-chemistryprize2020.pdf
How does CRISPR/Cas9 work?
In simple terms, Cas9 is the name of a protein that recognizes a specific sequence and cuts it. The recognition ability is due to a short piece of RNA (RNA is similar to DNA but slightly different in the type of sugar that makes it backbone) that can be attached to the Cas9 protein. This RNA is called a guide RNA and it basically guides the protein to the right place. In other words, the sequence (like DNA sequence) on the RNA, guides the RNA to a piece of DNA that has the same sequence (see DNA Sequencing part). Once the RNA finds the correct DNA sequence, the Cas9 protein (which is a nuclease) cuts the DNA. It is a natural system in bacteria. Scientisits figured out that if you put a piece of RNA with a specific sequence that you want changed and make it a part of the Cas9 protein, it will find the sequence on genomic DNA and cut it. With a few other modifications (a bit too complicated to explain), you can make big changes or very precise changes in the target genome. In fact, you can edit a single base if needed. Sickle Cell Anemia muation is one that has been corrected this way. Essentially, the way it can be used to treat diseaes involves two main strategies.
One involves taking cells out of our system that fights diseases (for example a cell type called T Cells), edit it (or engineer it) to allow the cells to target specific tumor associated antigens. This is also called immunotherapy (shown in Figure 1 below) and this method is widely known as CAR- T Cell (chimeric antigen receptor) therapy.
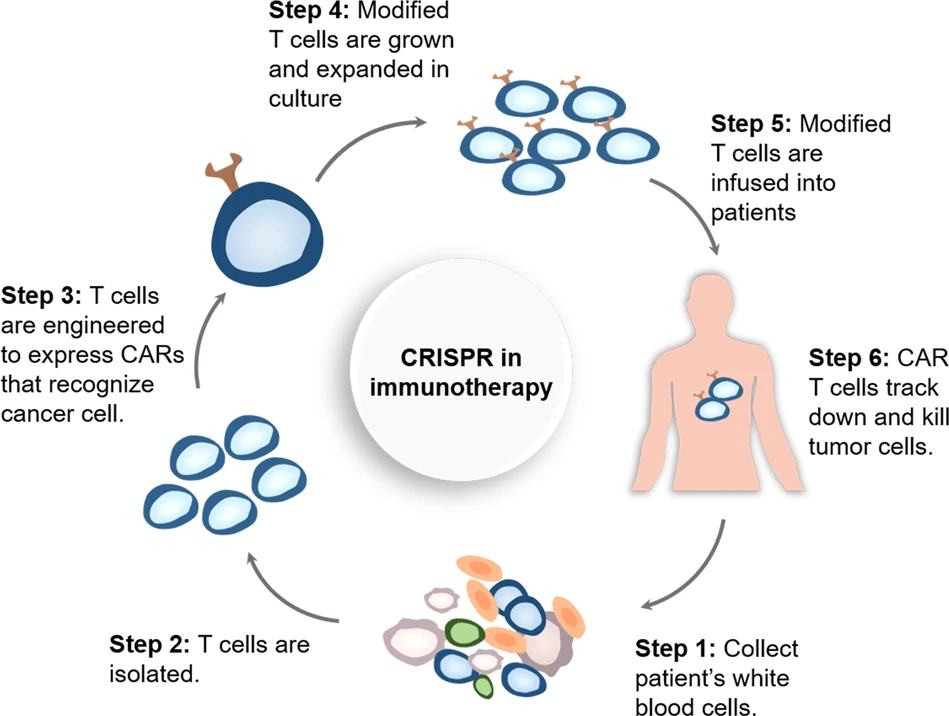
Many cancers have been fought with this approach. Such methods where cells are taken out of the body, modified and re-introduced back into the body are called ex-vivo methods.
Another way is to deliver the gene editing tools (like CRISPR) into live cell within the body. This is also being done for a variety of conditions and they are called in-vivo approaches. This can include putting the editing tools within viruses that are engineered to be targeted to specific cells within the body or using lipid nanoparticles to coat the tools. Both ex-vivo and in-vivo methods are summarized in Figure 2 below.
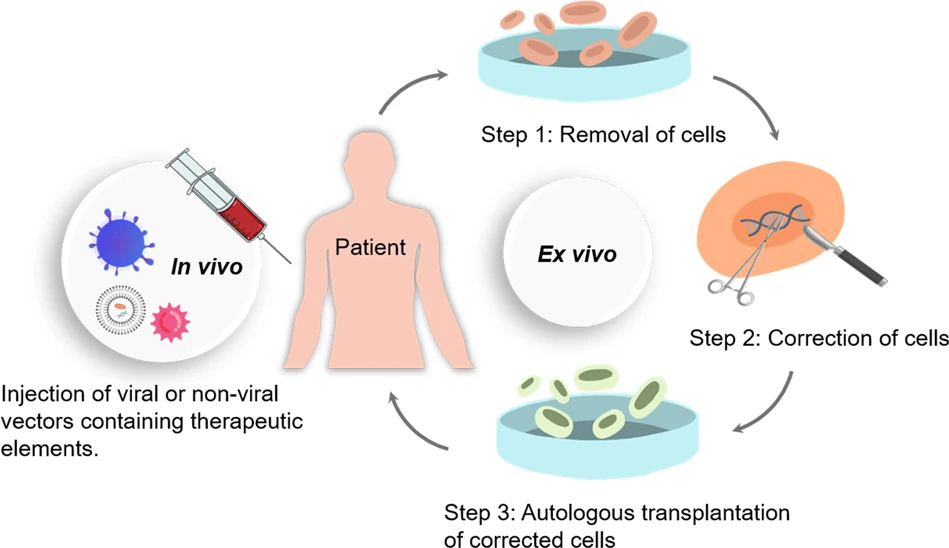
A good summary of these methods can be found in these links:
https://www.nature.com/articles/s41392-019-0089-y
https://www.nature.com/articles/s41581-022-00636-2
https://innovativegenomics.org/crispr-made-simple/crispr-health/
Both figures on this page were from the first link given above which is cited below:
i, H., Yang, Y., Hong, W. et al. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Sig Transduct Target Ther 5, 1 (2020).
https://doi.org/10.1038/s41392-019-0089-y
Both figures are used under the creative common license granted through Creative Commons CC BY